Web Tool Links Related to GPCR Study
General Molecular Biology Tools
33. MutSig2CV
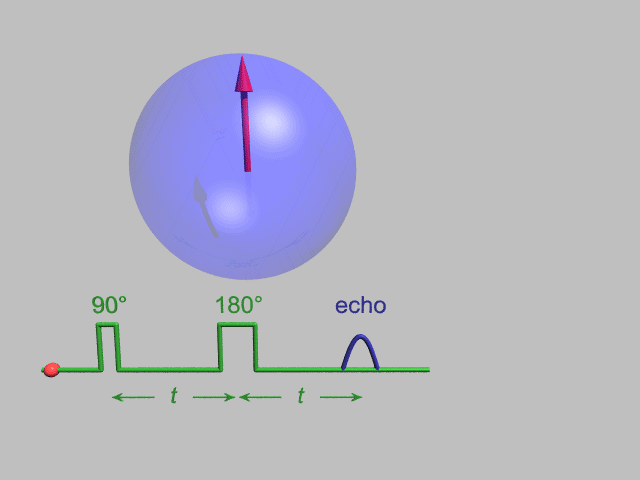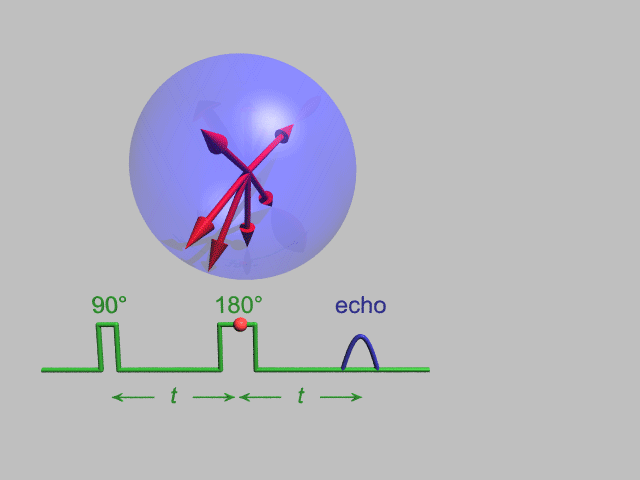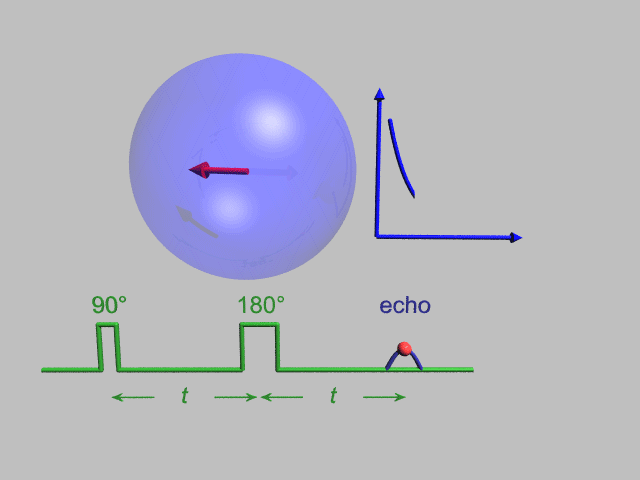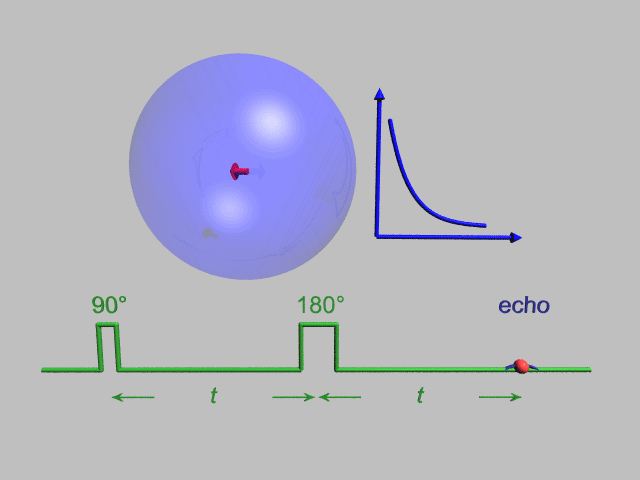
NMR and DEER
-
ANM Server
-
Basic nmr knowledge
-
Chemical fluoro
-
CPMG online fit software
-
DEER softwares
-
DEER Simulation Pronox
-
DESMOS WAVE FUNCTION
-
Library of NMR pulse
-
NMR course
-
NMRviewJ
-
Solvent shift and polarity
-
ProShift
-
Pulse sequence writing
-
The nmrPipeFit component
-
The basic NMR
-
Useful links from UC Davis
19. Protein chemical shifts
20. 1H,13C chemical shift prediction
21. NMR pulse sequence download
22. A pratical guide: Protein NMR
23. Chemical shift prediction
24. Protein chemical shift prediction
25. NMRFx (spectral processor)
26. Ad Bax--pulse sequence
27. NMR thermodynamics
Membrane Proteins (GPCR) Tools/Database
-
GPCR database
-
GPCR-I-TASSER
-
GPCR-SSFE2.0
-
GPCR Pharmacology
-
cDNA for GPCRs
-
GPCR-SAS (snakeplot)
-
GPCR interaction Network
-
MP structural dynamics gateway
-
Basic Pharm